
With the ROCR package you can also plot the ROC curve, lift curve and other model selection measures. ROC(classLabels,scores,Full=TRUE) Answer by erik for Calculate AUC in R?Īs mentioned by others, you can compute the AUC using the ROCR package. AUC is returned along with it's confidence interval and it is also mentioned on the graph. I usually use the function ROC from the DiagnosisMed package. Link to pROC Answer by George Dontas for Calculate AUC in R? The ROCR package will calculate the AUC among other statistics: auc.tmp data(aSAH) > # Syntax (response, predictor): > auc(aSAH$outcome, aSAH$s100b) Area under the curve: 0.7314 of "AUC: a Better Measure." seems to require knowing the class labels, and here is an example in MATLAB where I don't understand R(Actual = 1))īecause R (not to be confused with the R language) is defined a vector but used as a function? Answer by semaj for Calculate AUC in R? If you fit a different model (using, say, nlme::gls() that allows for nonhomogeneous error variances, then you will get unequal SEs in both the model summary and the emmeans results.Given a vector of scores and a vector of actual class labels, how do you calculate a single-number AUC metric for a binary classifier in the R language or in simple English? That is a consequence of the fact that the design is balanced (equal numers of observations in each cell) and the model assumption of a constant error SD. Notice that within each set of results above, the standard errors stay constant within a table.
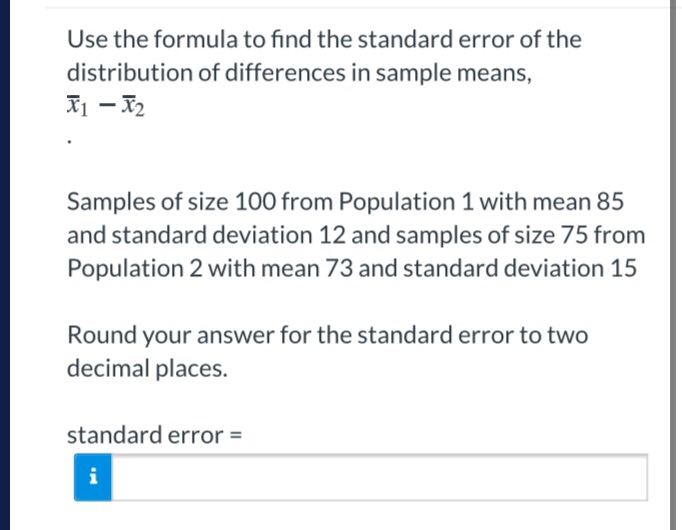
> contrast(emmeans(warp.lm, ~ wool*tension), interaction = "pairwise") For (1), note that the first result below matches the intercept, in both the estimate and the standard error: > emmeans(warp.lm, ~ wool * tension) We can observe these results in the output from emmeans() and its relatives.
The interaction coefficients are estimates of certain interaction contrasts (namely, differences of differences).In particular, they are comparisons of cell means where one factor is held constant while the other one changes.


First, the model itself: > getOption("contrasts")


 0 kommentar(er)
0 kommentar(er)
